This library have classes around the Chaos Game Representation for DNA sequence
The FCGR helps to visualize a k-mer distribution The FCGR of a sequence is an image showing the distribution of the
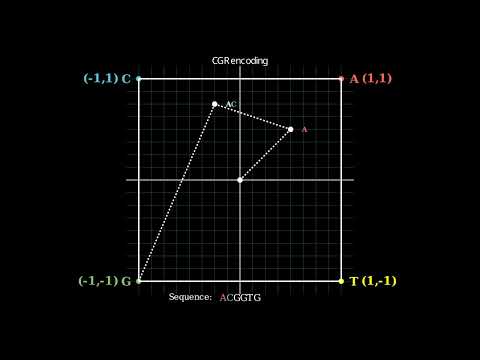
The position that a CGR.
Some examples of bacterial assemblies (see reference) are shown below.
The name of the species and the sample_id is in the title of each image (see an example with the first image). These images were
created using the 6-mers of each assembly and the class FCGR of this library.
 |
|---|
| 10 different species of bacteria represented by their FCGR (6-mers) |
pip install complexcgrto update to the latest version
pip install complexcgr --upgradefrom complexcgr import CGR
# Instantiate class CGR
cgr = CGR()
# encode a sequence
cgr.encode("ACGT")
# > CGRCoords(N=4, x=0.1875, y=-0.5625)
# recover a sequence from CGR coordinates
cgr.decode(N=4,x=0.1875,y=-0.5625)
# > "ACGT"Input for FCGR only accept sequences in
import random; random.seed(42)
from complexcgr import FCGR
# set the k-mer
fcgr = FCGR(k=8) # (256x256) array
# Generate a random sequence without T's
seq = "".join(random.choice("ACG") for _ in range(300_000))
chaos = fcgr(seq) # an array with the frequencies of each k-mer
fcgr.plot(chaos) |
|---|
| FCGR representation for a sequence without T's |
You can save the image with
fcgr.save_img(chaos, path="img/ACG.jpg")Formats allowed are defined by PIL.
You can also generate the image in 16 (or more bits), to avoid losing information of k-mer frequencies
# Generate image in 16-bits (default is 8-bits)
fcgr = FCGR(k=8, bits=16) # (256x256) array. When using plot() it will be rescaled to [0,65535] colors# Generate a random sequence without T's and lots of N's
seq = "".join(random.choice("ACGN") for _ in range(300_000))
chaos = fcgr(seq) # an array with the probabilities of each k-mer
fcgr.plot(chaos) |
|---|
| FCGR representation for a sequence without T's and lots of N's |
from complexcgr import iCGR
# Instantiate class CGR
icgr = iCGR()
# encode a sequence
icgr.encode("ACGT")
# > CGRCoords(N=4, x=3, y=-9)
# recover a sequence from CGR coordinates
icgr.decode(N=4,x=3,y=-9)
# > "ACGT"from complexcgr import ComplexCGR
# Instantiate class CGR
ccgr = ComplexCGR()
# encode a sequence
ccgr.encode("ACGT")
# > CGRCoords(k=228,N=4)
# recover a sequence from ComplexCGR coordinates
ccgr.decode(k=228,N=4)
# > "ACGT"Input for FCGR only accept sequences in
import random; random.seed(42)
from complexcgr import FCGR
# set the k-mer desired
cfcgr = ComplexFCGR(k=8) # 8-mers
# Generate a random sequence without T's
seq = "".join(random.choice("ACG") for _ in range(300_000))
fig = cfcgr(seq) |
|---|
| ComplexFCGR representation for a sequence without T's |
You can save the image with
cfcgr.save(fig, path="img/ACG-ComplexCGR.png")Currently the plot must be saved as png
Count k-mers could be the bottleneck for large sequences (> 100000 bp).
Note that the class FCGR (and ComplexCGR) has implemented a naive approach to count k-mers, this is intended since in practice state-of-the-art tools like KMC or Jellyfish are used to count k-mers very efficiently.
We provide the class FCGRKmc, that receives as input the file generated by the following pipeline using KMC3
Make sure to have kmc installed. One recommended way is to create a conda environment and install it there
kmer_size=6
input="path/to/sequence.fa"
output="path/to/count-kmers.txt"
mkdir -p tmp-kmc
kmc -v -k$kmer_size -m4 -sm -ci0 -cs100000 -b -t4 -fa $input $input "tmp-kmc"
kmc_tools -t4 -v transform $input dump $output
rm -r $input.kmc_pre $input.kmc_sufthe output file path/to/count-kmers.txt can be used with FCGRKmc
from complexcgr import FCGRKmc
kmer = 6
fcgr = FCGRKmc(kmer)
arr = fcgr("path/to/count-kmers.txt") # k-mer counts ordered in a matrix of 2^k x 2^k
# to visualize the distribution of k-mers.
# Frequencies are scaled between [min, max] values.
# White color corresponds to the minimum value of frequency
# Black color corresponds to the maximum value of frequency
fcgr.plot(arr)
# Save it with numpy
import numpy as np
np.save("path_save/fcgr.npy",arr)CGR encoding
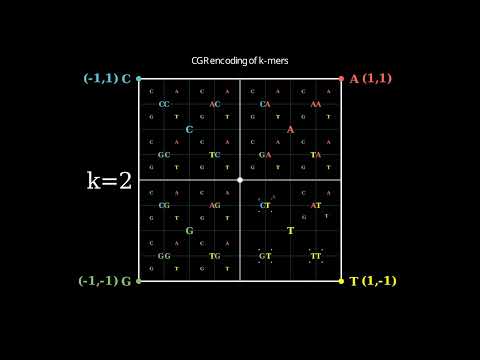
CGR encoding of all k-mers
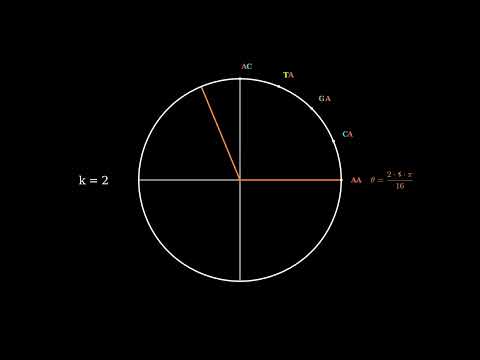
ComplexCGR encoding
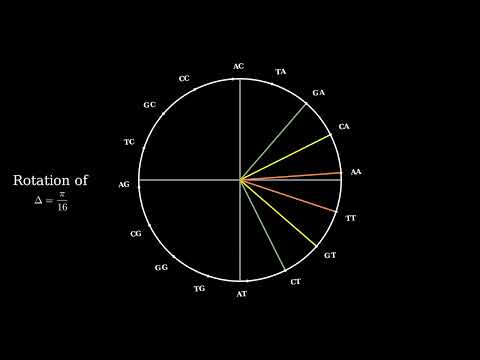
ComplexCGR and Symmetry
version 0.8.0:
A list of available classes and functionalities are listed below:
Encoders
The encoders are functions that map a sequence CGR, iCGR, and ComplexCGR.
CGR Chaos Game Representation: encodes a DNA sequence in 3 numbers
- encode a sequence.
- recover a sequence from a CGR encoding.
iCGR integer CGR: encodes a DNA sequence in 3 integers
CGR Chaos Game Representation: encodes a DNA sequence in 3 numbers
- encode a sequence.
- recover a sequence from a CGR encoding.
iCGR integer CGR: encodes a DNA sequence in 3 integers
- encode a sequence
- recover a sequence from an iCGR encoding
ComplexCGR: encodes a DNA sequence in 2 integers
- encode a sequence
- recover a sequence from a ComplexCGR encoding
- plot sequence of ComplexCGR encodings
Image for distribution of k-mers
-
FCGRFrequency Matrix CGR: representation as an image for k-mer representativity, based on CGR.- generates FCGR from an arbitrary n-long sequence.
- plot FCGR.
- save FCGR generated.
- save FCGR in different bits.
-
FCGRKmcSame asFCGRbut receives as input the file with k-mer counts generated with KMC -
ComplexFCGR: Frequency ComplexCGR: representation as an image (circle) for k-mer representativity, based on ComplexCGR.- generates ComplexFCGR from an arbitrary n-long sequence.
- plot ComplexFCGR.
- save ComplexFCGR generated.
-
PercentileFCGR: -
SpacedFCGR: Create FCGR from spaced-mers
complexcgr is developed by Jorge Avila Cartes