-
Notifications
You must be signed in to change notification settings - Fork 101
REL: Release 1.0.0 #243
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
REL: Release 1.0.0 #243
Conversation
Add encrypted travis API keys to the .tavis.yml file for test.pypi and pypi.
Change username in .travis.yml repo git URL to be the organisation name 'TIA-LAB' instead of the GitHub user 'tialab'.
Apply python Black formatting to all files, mostly affects files generated with Cookiecutter.
* Add code coverage XML report generation via pytest-cov by adding test runner options in tox.ini file. * Create .coveragerc file for pytest-cov using codecov defaults. * Add report upload to codecov and deepsource services by adding after-sucess section to .travis.yml file.
* BUG: Fix .travis.yml indentation Fix indented lines in the after_sucess section of the .travis.yml configuration file. * BUG: Add missing space in travis configuration Add missing space to deepsource test upload line in .travis.yml configuration.
DEV: Add feature to collect slide meta data Merge pull request #1 from develop Add Feature slide-info - Add WSIReader class to read whole slide images. Currently this class is using openslide and therefore can only read file formats supported by openslide - Add slide_info function which returns slide meta information - Add TIAMultiProcess decorator to run functions using multiple cores. - Add misc to write miscellaneous small functions repeatedly required for use in the repo.
Merge pull request #5 from docs-readthedocs-config Create Read the Docs configuration file (.readthedocs.yml) specifying to use conda setup to create environment using requiremetns.conda.yml. - Add Conda Forge OpenSlide To Requirements - Add OpenSlide to conda requirements file. - Add Pixman < 0.38.0 (0.38.0 breaks OpenSlide). - Removed version numbers from others so that latest is used. - Pin Setup Tools Verions <= 45.1 - Pin OpenCV Version >4.0 In Conda Requirements File - Add requirements.txt and requirements.win64.conda.yml - Modify setup.py to add dependencies - Make version numbers for various packages in requirements and conda yaml consistent. Pin version numbers if necessary. Co-authored-by: shaneahmed <sa>
- Add feature read_region to WSIReader class in wsireader.py - Read region allows to read an image region from a whole slide image using openslide-python api. This image can be saved to disk or shown via the default image viewer. - Update README.rst -Update README.rst to update `pip install -r requirements.txt` instead of `pip install requirements_dev.txt` Co-authored-by: shaneahmed <sa> Co-authored-by: John Pocock <[email protected]>
Add release notes for version 0.2.0 in HISTROY.rst.
- Change files shared between GitHub/PyPI and Sphinx/ReadTheDocs from ReStructured Test to Markdown. These are readme, histroy, authors. - Add recommonmark in new requiremenst.dev.conda.yml file - Add symlinks in docs/ to root readme files for reade, authors, history.
Change Read The Docs configuration YAML to use the development conda environment file.
- Change setup.py to use .md files instead of .rst - Add long_description_content_type
Change .rst files to .md
- Change version strings in setup.py and tiatoolbox/__init__.py - Change bum2version rules in setup.cfg for look for double quotes
Fix slide_info mode argument through cli
Bring version number change from master which fixed small deployment configuration issue.
Mainly concern with making the doc looks better. - [x] Automate API generation instead of manual linkage. - [x] Add back intro page or swap header names to make it easier for new python user navigation. Co-authored-by: Dang Vu <[email protected]>
- Add ER status prediction to SlideGraph+ notebooks Co-authored-by: Wenqi Lu <[email protected]> Co-authored-by: Dang Vu <[email protected]>
- Separate out readthedocs requirements to avoid python 3.7 requirements errors.
- Add detection of pixman version with a warning for 0.38. Currently works: - In a conda env on all platforms. - When OpenSlide (and pixman as a dependency) is installed with homebrew on all platforms (macOS, and linux). - When OpenSlide (and pixman as a dependency) is installed with macports on macOS (needs testing). - Add detection of conda environment. - Add detection of notebook environments, CoLab, and Kaggle (e.g. to change default parameters or display a message). - Add detection of CoLab with GPU. - Merged coverage configuration into global config (pyproject.toml) - Excluded detection from coverage as it is not possible (or easy to do properly) to test on travis. - Added simple network connection test via a ping to our server. # Pixman Warnings ## Conda Env ``` $ python >>> from tiatoolbox.wsicore import wsireader |2021-12-02|14:14:19.971| [WARNING] It looks like you are using Pixman version 0.38 (via conda). This version is known to cause issues with OpenSlide. Please consider upgrading to Pixman versi on 0.39 or later. You may be able do this with the command: conda install -c conda-forge pixman ">=0.39" ``` ## Ubuntu (installed via apt) ``` $ python >>> from tiatoolbox.wsicore import wsireader |2021-12-02|14:14:19.971| [WARNING] It looks like you are using Pixman version 0.38 (via dpkg). This version is known to cause issues with OpenSlide. Please consider upgrading to Pixman versi on 0.39 or later. To fix this you may need to set up an anaconda environment with pixman >=0.39 or install pixman >=0.39 from source. See the tiatoolbox documentation for more information on setting up a conda environment. Instructions to compile from source can be found at the GitLab mirror here: https://gitlab.freedesktop.org/pixman/pixman/-/blob/master/INSTALL ``` ## macOS Homebrew ``` $ python >>> from tiatoolbox.wsicore import WSIReader |2021-12-02|14:14:19.971| [WARNING] It looks like you are using Pixman version 0.38 (via brew). This version is known to cause issues with OpenSlide. Please consider upgrading to Pixman versi on 0.39 or later. You may be able do this with the command: brew upgrade pixman. ``` # Detection Examples ## Detecting CoLab ```python >>> from tiatoolbox.utils import detection >>> detection.running_on_colab() False ``` ## Detecting Notebook ```python >>> from tiatoolbox.utils import detection >>> detection.is_notebook() False ``` ## Detecting Conda ```python >>> from tiatoolbox.utils import detection >>> detection.in_conda_env() True ``` ## Running on Travis ```python >>> from tiatoolbox.utils import detection >>> detection.running_on_travis() False ``` ## Detecting Interactive Returns True for interactive environments such as notebook, python REPL, or PyCharm console etc. ```python >>> from tiatoolbox.utils import detection >>> detection.is_interactive() True ``` Co-authored-by: John Pocock <[email protected]> Co-authored-by: Shan Raza <[email protected]>
- Rename: - FeatureExtractor to DeepFeatureExtractor - CNNExtractor to CNNBackbone Co-authored-by: Dang Vu <[email protected]> Co-authored-by: Shan Raza <[email protected]>
The aim of this PR (which is a branch off of tidying wsireader.py) aims to make imports only happen as needed where possible. This means that some dependencies can be avoided if they are not required. On demand imports so far: - glymur (and openjpeg) - defusedxml Co-authored-by: John Pocock <[email protected]> Co-authored-by: ShanRaza <[email protected]>
- Add citation to the git Co-authored-by: Shan Raza <[email protected]> Co-authored-by: John Pocock <[email protected]>
…#229) - Use normalize (US Spellings) instead of normalise (UK Spellings) Co-authored-by: Shan Raza <[email protected]> Co-authored-by: Simon Graham <[email protected]>
- Add example notebook for the multi-task segmentor. Co-authored-by: Adam Shephard <[email protected]>
- Update the whole slide image used in the multi-task segmentor notebook. Co-authored-by: Adam Shephard <[email protected]>
- Update links to Colab, Git and Kaggle Co-authored-by: Shan Raza <[email protected]>
…#209) - Allow users to provide mpp and objective power values when opening an image / initialising a reader if they know the value. Co-authored-by: John Pocock <[email protected]>
…#217) - Improve performance of mask based patch extraction. In the current implementation, the `filter_coordinates` function is used to filter out generated coordinates based on the input mask. This is done by reading the mask region from `mask_reader` object (which is a VirtualWSI) at the same resolution and region requested in the patch extraction. Therefore, this process can take very long specially if the whole slide mask region is big and user requests to extract small patches from highest resolution (not to mention the increase in time due to overlap in patch extraction). For example, if you want extract 256x256 patches at 40x from the below WSI, it might take up to **10 minutes** for checking filter the coordinates   In this PR, I have introduced `filter_coordinates_fast` which uses the overview of `mask_reader` object (a numpy array) at a fixed-low resolution (mpp=8 or power=1.25) to speed of the region mask reading process and hence improve the speed of coordinate filtering and whole patch extraction processes. For example, the above image takes about **0.5 seconds** (at least **1,000x improvement in speed** in comparison to previous method). 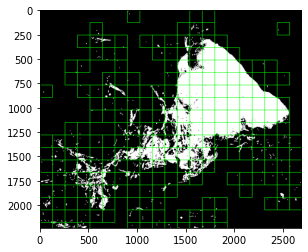 To be able to do this, I also introduce a method to `WSIReader` class that converts resolution from one unit to all other units, because the `filter_coordinates_fast` expects its input mask resolution to be in `mpp`, `power`, or `baseline` units. - [x] Implement fast coordinate filtering function - [x] Implement resolution conversion function in wsireader - [x] Writing tests - [x] Improve coverage Co-authored-by: Mostafa Jahanifar <[email protected]> Co-authored-by: Dang Vu <[email protected]> Co-authored-by: John Pocock <[email protected]>
- Update Stain normalization notebook Co-authored-by: Shan Raza <[email protected]>
- Add Missing Cell for Semantic Segmentation Notebook
- Update titles on Colab for notebooks. Co-authored-by: Adam Shephard <[email protected]>
- Update examples/05-patch-prediction
- Fix readthedocs errors by updating the metadata in notebooks - Fix examples/08-nucleus-instance-segmentation Co-authored-by: Shan Raza <[email protected]>
- Change PDF Build Engine to fix Unicode errors. Co-authored-by: Dang Vu <[email protected]>
- Add HoVer-Net CoNSeP and MoNuSAC models Co-authored-by: Simon Graham <[email protected]>
- Include all pretrained model info in `pretrained.rst`.
- Fix Kaggle error with bash Co-authored-by: Shan Raza <[email protected]>
### Major Updates and Feature Improvements - Adds nucleus instance segmentation base class - Adds [HoVerNet](https://www.sciencedirect.com/science/article/abs/pii/S1361841519301045) architecture - Adds multi-task segmentor [HoVerNet+](https://arxiv.org/abs/2108.13904) model - Adds <a href="https://www.thelancet.com/journals/landig/article/PIIS2589-7500(2100180-1/fulltext">IDaRS</a> pipeline - Adds [SlideGraph](https://arxiv.org/abs/2110.06042) pipeline - Adds PCam patch classification models - Adds support for stain augmentation feature - Adds classes and functions under `tiatoolbox.tools.graph` to enable construction of graphs in a format which can be used with PyG (PyTorch Geometric). - Add classes which act as a mutable mapping (dictionary like) structure and enables efficient management of annotations. (#135) - Adds example notebook for adding advanced models - Adds classes which can generate zoomify tiles from a WSIReader object. - Adds WSI viewer using Zoomify/WSIReader API (#212) - Adds README to example page for clarity - Adds support to override or specify mpp and power ### Changes to API - Replaces `models.controller` API with `models.engine` - Replaces `CNNPatchPredictor` with `PatchPredictor` ### Bug Fixes and Other Changes - Fixes Fix `filter_coordinates` read wrong resolutions for patch extraction - For `PatchPredictor` - `ioconfig` will supersede everything - if `ioconfig` is not provided - If `model` is pretrained (defined in `pretrained_model.yaml` ) - Use the yaml ioconfig - Any other input patch reading arguments will overwrite the yaml ioconfig (at the same keyword). - If `model` is not defined, all input patch reading arguments must be provided else exception will be thrown. - Improves performance of mask based patch extraction ### Development related changes - Improve tests performance for Travis runs - Adds feature detection mechanism to detect the platform and installed packages etc. - On demand imports for some libraries for performance - Improves performance of mask based patch extraction Co-authored-by: Shan Raza <[email protected]>
|
Check out this pull request on See visual diffs & provide feedback on Jupyter Notebooks. Powered by ReviewNB |
Codecov Report
@@ Coverage Diff @@
## master #243 +/- ##
==========================================
+ Coverage 99.79% 99.81% +0.02%
==========================================
Files 40 53 +13
Lines 2959 4950 +1991
Branches 492 827 +335
==========================================
+ Hits 2953 4941 +1988
- Misses 1 2 +1
- Partials 5 7 +2
Continue to review full report at Codecov.
|
- Fix README conda link
Major Updates and Feature Improvements
tiatoolbox.tools.graphto enable construction of graphs in a format which can be used with PyG (PyTorch Geometric).Changes to API
models.controllerAPI withmodels.engineCNNPatchPredictorwithPatchPredictorBug Fixes and Other Changes
filter_coordinatesread wrong resolutions for patch extractionPatchPredictorioconfigwill supersede everythingioconfigis not providedmodelis pretrained (defined inpretrained_model.yaml)modelis not defined, all input patch reading arguments must be provided else exception will be thrown.Development related changes